This page describes the three-letter code used for residue names in files relevant to the GLYCAM force fields. For information on other nomenclatures recognized by GLYCAM-Web, please click here.
Current Carbohydrate Naming Convention in GLYCAM_04 and GLYCAM_06
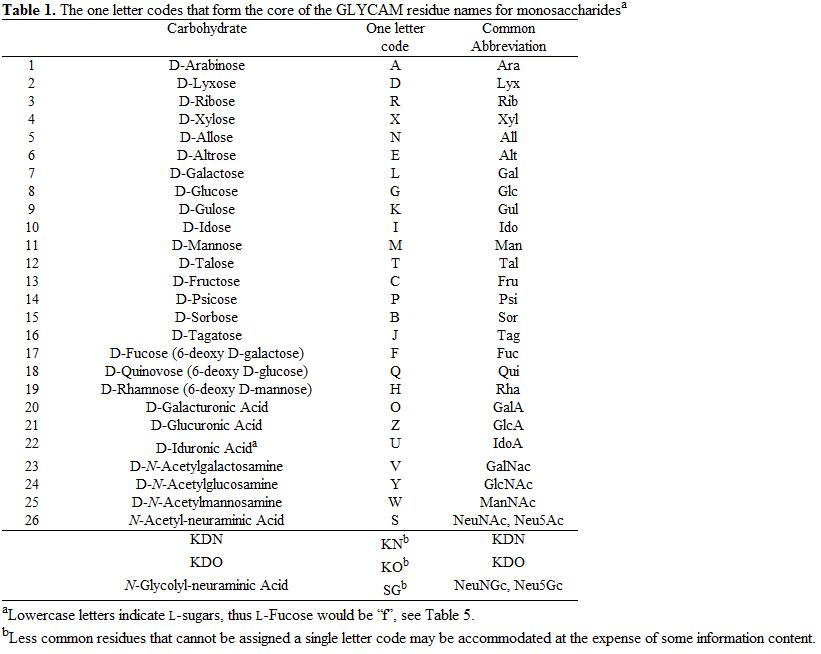
In order to develop a versatile set of 3-D structural modeling tools for carbohydrates, glycoproteins and carbohydrate-protein complexes (http://glycam.ccrc.uga.edu), we have introduced the following standards for one-letter (Table 1) and three-letter (Tables 2-5) codes for monosaccharides. There are only 26 possible one-letter codes, and they have been assigned to all of the hexoses and pentoses, as well as to the common 6‑deoxy‑ and 2‑N‑acetamido derivatives, uronic acids and sialic acid. Where possible, the letter is taken from the first letter of the monosaccharide name, however, (A = Ara, F=Fuc, G=Glc, I=Ido, M=Man, P=Psi, Q=Qui, R = Rib, T=Tal, X=Xyl). L was assigned to Gal for alliterative reasons, though it could have been used for the much less common Lyx.
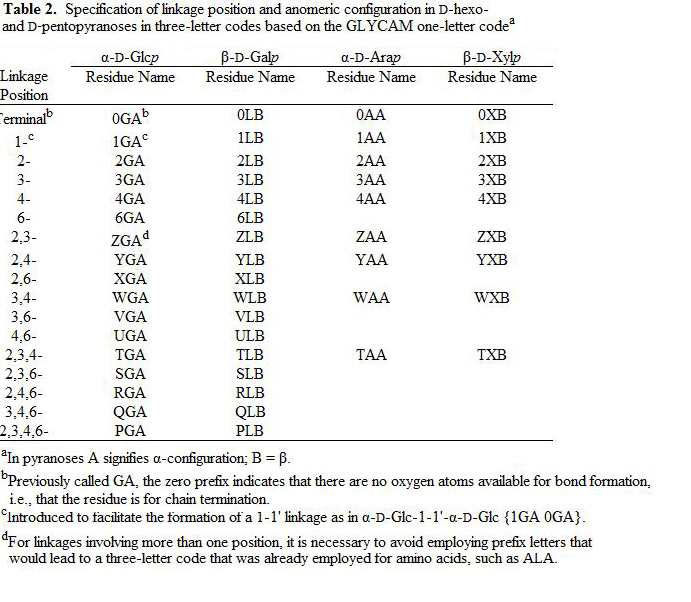
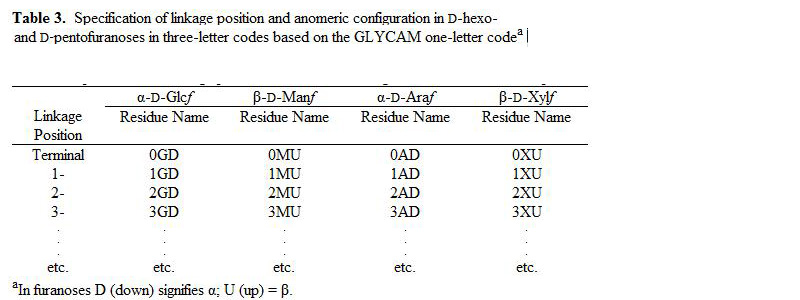
In order to incorporate carbohydrates in a standardized way into modeling programs, as well as to provide a standard for X-ray and NMR protein database files (pdb), we have developed the following three-letter code nomenclature. The restriction to three letters is based on standards imposed on protein database (pdb) files by the RCSB PDB Advisory Committee (http://www.rcsb.org/pdb/pdbac.html), and for the practical reason that all current modeling and experimental software has been developed to read three-letter codes, historically for amino acids.
Special Notes About Alternate Residue Codes
The GLYCAM06 parameter set has begun to expand beyond the set of residues treated by the three-letter naming convention for residues. Here, we introduce three alternate classes of residue codes: four-letter codes, three-letter codes with non-standard naming, and codes where the anomeric configuration is specified by case in the third character.
In general, to understand any particular code, it is necessary to consult documentation associated with the release of the residue structural data.
Four-Letter Codes
Occasionally, it is desirable to have more than one set of structures available for a given residue. For example, iduronate (IdoA, code ‘U’) exists in nature in three ring conformations: 1C4, 4C1 and 2SO. To differentiate them, a fourth character (‘1’, ‘2’ or ‘3’) is added to the three-letter code. So, 4uA is available as 4uA1, 4uA2 and 4uA3 in our parameter set.
Non-Standard Three-Letter Naming
When a residue is not covered at all in the parameter set, it might be assigned almost anything. If necessary to avoid confusion, it might be assigned an all-numerical code, for example ‘001’ or ‘999’. These are defined in files associated with their release.
Specification of Anomeric Configuration in Third Character
In some codes, for example those for GlcNS, the anomeric configuration (alpha or beta) is specified by the case in the third character, with alpha as capital and beta as lower case. For example, 0YN is alpha-D-Glucosamine, and 0Yn is beta.
Combinations of Alternate Coding
Some codes combine one or more element. For example, protonated alpha-D-Glucosamine is 0YNP.
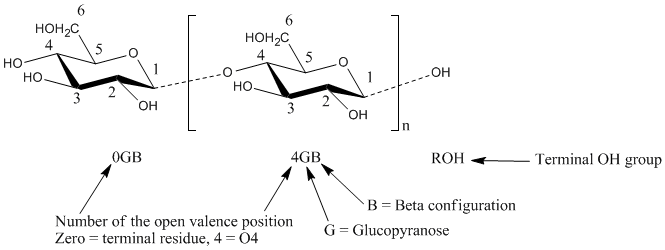
Figure 1. Illustration of the GLYCAM naming convention applied to cellobiose.
Using three letters (Figure 1, Tables1-5), the present system encodes the following content: carbohydrate residue name (Glc, Gal, etc.), ring form (pyranosyl or furanosyl), anomeric configuration (α or β), enantiomeric form (D or L) and occupied linkage positions (2-, 2,3-, 2,4,6-, etc.). Incorporation of linkage position is a particularly useful addition, since, unlike amino acids, the linkage cannot otherwise be inferred from the monosaccharide name. Further, the three-letter codes were chosen to be orthogonal to those currently employed for amino acids. Please see Figure 1 for a graphical illustration of the naming convention.