Overview
This document describes the monosaccharide representation scheme used by GMML in its monosaccharide identification. It is essentially a monosaccharide shorthand for humans to use when needing to translate a visual representation of a cyclic monosaccharide into its name. Since it is also linearizable, it is similarly useful in computer programs.
Rationales
1. Ease of communication
It is useful to have a method for representing monosaccharides that is:
- Simpler than a typical atomistic representation such as IUPAC or SMILES.
- Represents 3D geometry well, but can be written in plain text, unlike a Fischer or Haworth projection.
- Easy for someone with basic chemistry training to produce and understand.
- Can be linearized for ease of use in computing algorithms.
2. Focus on the monosaccharide ring
In a glycobiological context, the recognition of shapes, for binding or enzymatic activity, is of considerable significance. Furthermore, these recognition events often involve the monosaccharide ring. If the carbon chain is extended in either direction away from the ring, the official naming changes in a way to make it nearly impossible to recognize similarities in this important recognition region. This code concerns itself first with rings, then with what is attached to them
Computationally speaking, it is useful to first find all the cycles in a structure and then build a representation based on the cycle. Of course, this doesn’t work for linear monosaccharides, but they have not been shown to be as biologically important as the cyclic ones.
3. One-to-one mapping between structure and code
To be able to unambiguously identify monosaccharides, it is necessary that the representation be able to encode one and only one 3D structure.
4. Tolerance of ambiguity in the 3D structure
3D structures are sometimes flawed. And, if one needs to use a representation like this, it is very likely that a potential flaw in a structure is the reason for that. It is useful that the representation not break if there are missing or extra atoms, etc.
Caveats
This documentation assumes that you already know something about monosaccharide structure.
Currently, the scheme, and associated identification code, treats only cyclic monosaccharides whose rings contain 5 (furanose) or 6 (pyranose) members. It could be extended to linear monosaccharides or other ring types.
The representation scheme might undergo additions or changes if unforeseen needs arise. Every attempt will be made to minimize changes and to ensure that additions do not add confusion. If changes are made, whenever possible, they will not cause earlier versions to be unreadable or indeterminate.
The name “GlyCode” is not associated with any commercial, software or organizational entities. It’s just a name.
Similar Codes
Most people who have coded glycan recognition sofware have used some sort of very similar scheme. For example, the code used in the PDB monosaccharide recognition software at glycosciences.de, uses a series of numbers to represent ring size and to indicate that the substituents are ‘down’ or ‘up’ at each ring position (PMID: 15010309). This representation is certainly simple for computers to use, but is less meaningful for humans. Our identification scheme is quite similar to theirs in several ways, but we hope to have improved upon it.
Representation Scheme
The representation scheme is based on the standard nomenclature of the common monosaccharides. It has a format that chemists will find familiar. It eases the translation between 3D structural representations and look-up tables.
In the figure below, a 3D image of alpha-D-glucopyranose (leftmost) can be rendered as text. The text can be easily compared to a lookup table. The table in the image comes from the book Conformation of Carbohydrates.
Structure Basics – Simple Monosaccharides
Encoding a structure requires six steps:
- Using visualization software, or your imagination, etc., orient the ring so that the ring oxygen is at your right and the anomeric center is towards you, as shown here:
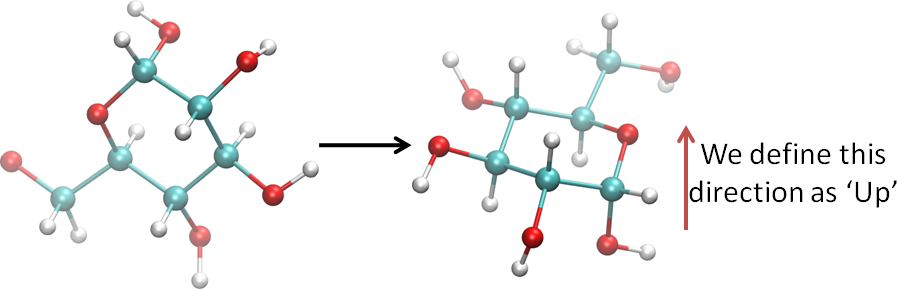
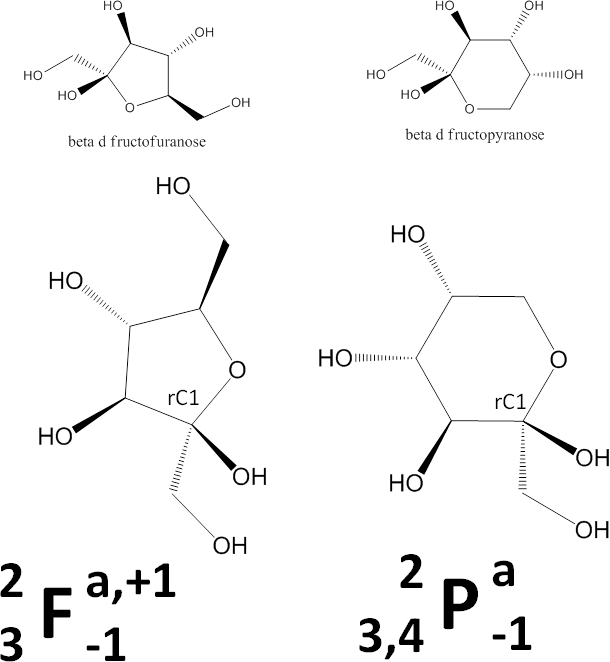
- Determine whether the ring is a pyranose (P) or furanose (F). This image shows chemical schemes of beta-D-fructofuranose (left) and beta-D-fructopyranose (right).
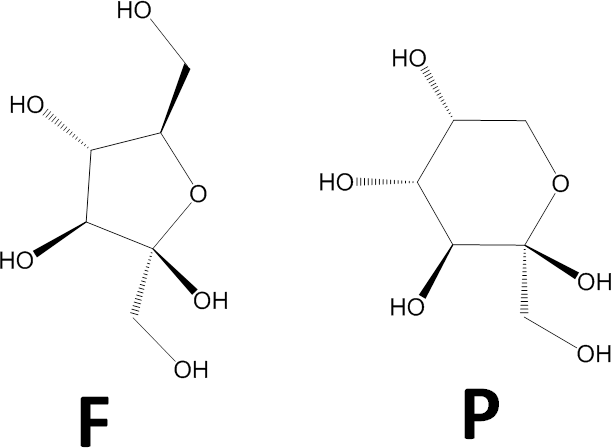
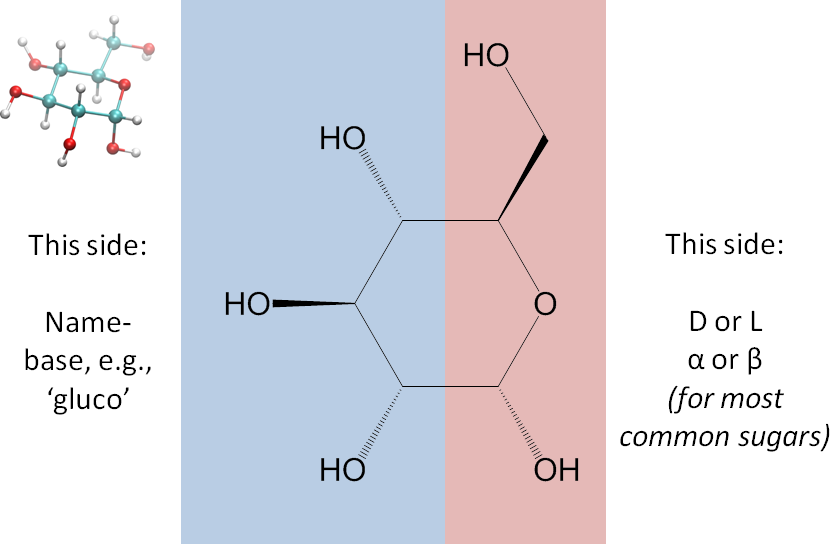
This letter, P or F, will be the central feature in the representation. - Divide the monosaccharide as shown in the image below. We dividing it this way so that on the right side are the parts that in the common sugars determine whether the monosaccharide is alpha or beta and D or L, and on the left side are the parts that generally give rise to its name (gluco, manno, talo, etc.). A ball-and-stick 3D representation is included in the image for your convenience.
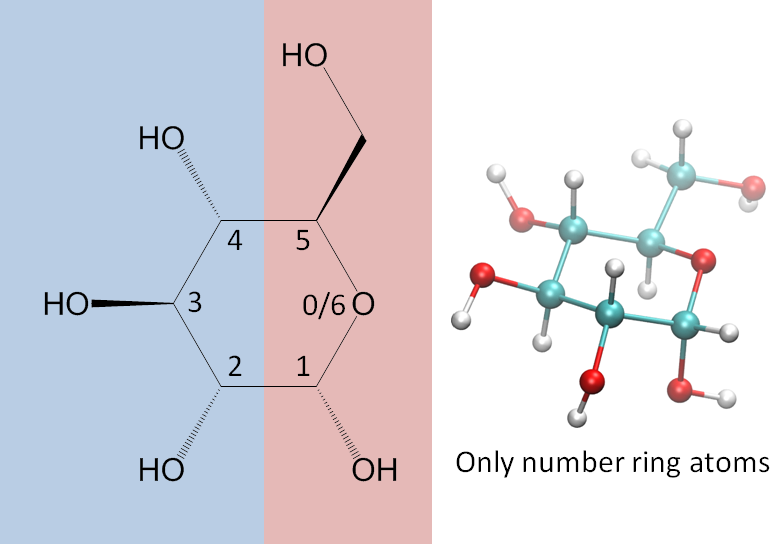
- Next, number the ring atoms. NOTE!! Most chemists have been taught to number the longest carbon chain. That is great to do when assigning IUPAC names, but isn’t the thing to do here. Our concern is with the ring, so number only the ring atoms. Note that the ring oxygen can be considered position 0 (zero) or the last one (here, 6), depending on the context.
- Determine the orientations of heavy atoms at each ring position.
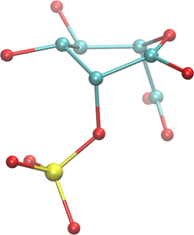 In the case of a heavily-distorted ring, this might not be a trivial thing to do. The following brief procedure outlines an easy method for doing so when visualizing a structure. But, it can easily be rendered as a computer algorithm, and it is the one used in GEMS/GMML’s Sugar ID code. In this example, we will consider the highly distorted structure at right. We will refer to ring atoms as rC or rO for clarity.
In the case of a heavily-distorted ring, this might not be a trivial thing to do. The following brief procedure outlines an easy method for doing so when visualizing a structure. But, it can easily be rendered as a computer algorithm, and it is the one used in GEMS/GMML’s Sugar ID code. In this example, we will consider the highly distorted structure at right. We will refer to ring atoms as rC or rO for clarity.
-
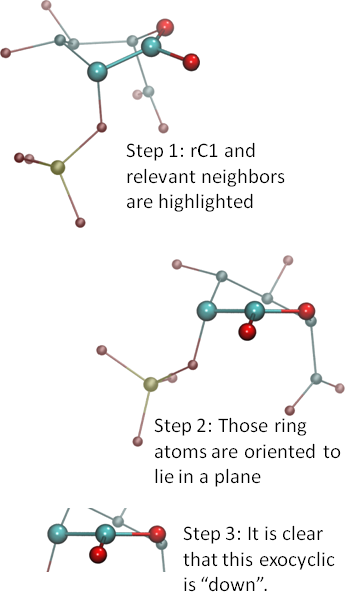 Consider the ring atom for which you need orientation information and the ring atoms just before and after it. In the example at right, we are considering ring carbon one (rC1), so that we can determine whether the oxygen attached to it is ‘up’ or ‘down’ or ‘in the plane’.
Consider the ring atom for which you need orientation information and the ring atoms just before and after it. In the example at right, we are considering ring carbon one (rC1), so that we can determine whether the oxygen attached to it is ‘up’ or ‘down’ or ‘in the plane’. - Orient the three atoms so that the ring atom numbers increase from right to left. Here, we are numbering the ring oxygen (rO) as rO0, so that the numbers go 0-1-2 from right to left. Once you have the atoms in the proper order, reorient them so that those three ring atoms lie in a plane. See Step 2 in the image. Note that the exocyclic should be pointing towards you unless the structure is very unusual.
- If the exocyclic is above the plane, then it is ‘up’. If below the plane, ‘down’. Exocyclics that are perfectly in the same plane, or nearly so, suggest an unusual electronic structure, such as unsaturation. But, whatever the case, consider them to be ‘in the plane’ and neither up nor down.
-
 Use the positions determined in the previous step to finish encoding the structure using the following rules. See also the color-coded image at right.
Use the positions determined in the previous step to finish encoding the structure using the following rules. See also the color-coded image at right.
- Recall the ring division in Step 3. Add the positions of exocyclic hydroxyls that were on the left in Step 3 to the left side of the P or F as determined in Step 2. Make them be super-scripted if they are ‘up’ and sub-scripted if they are ‘down’.
- Use only numbers if the attached atoms are hydroxyl groups. The next section addresses derivatives.
- If an exocyclic is ‘in the plane’ (neither up nor down), do not super- or sub-script it. There are examples of this below.
- Add positions of the atoms at positions flanking the ring oxygen to the right side of the P or F.
- Since the anomeric center is very important, we note its position as a lower-case ‘a’. We will see the use of an upper-case ‘A’ below.
- Any heavy atom extending from the non-anomeric side of the ring oxygen is given a positive number. Here, the carbon with absolute number 6 extends from the non-anomeric side. Since it has the expected hydroxyl attached to it, it gets only the numerical representation ‘+1’. It is super-scripted because it is in an ‘up’ position relative to its ring-atom neighbors.
- Any heavy atom extending from the anomeric side, and which is not the anomeric oxygen (e.g., as with a keto sugar), is designated by a negative number. Examples of this are given below.
- If needed for complex exocyclics, chirality can be expressed using standard methods. Examples of this are also given below.
- Recall the ring division in Step 3. Add the positions of exocyclic hydroxyls that were on the left in Step 3 to the left side of the P or F as determined in Step 2. Make them be super-scripted if they are ‘up’ and sub-scripted if they are ‘down’.
Ketose Examples
To illustrate the use of the negative numbers mentioned in Step 6.2.3, we present representations for the furano and pyrano forms of beta-D-fructose. Other uses of negative numbers occur below.
Writing Linear Representations
 The monosaccharide we have been considering so far is alpha-D-glucopyranose. A scheme of its chemical structure and its glycode are shown at right. A linearized form is:
The monosaccharide we have been considering so far is alpha-D-glucopyranose. A scheme of its chemical structure and its glycode are shown at right. A linearized form is:
_2^3_4P^+1_a
Here, subscripts are preceded by an underscore and superscripts are preceded by a caret.
Linearization versions can vary a little, provided that they are not confusing and that the variants all obviously have the same meaning. For example, the programmers of the Sugar ID preferred that each position be given a super- or sub-script notation and that the ring positions be listed in order. But, the following would also be acceptable:
_2,4^3P_a^+1
To indicate that an entity is neither super- nor sub-scripted, use parentheses around it. For example, beta-D-abequopyranose (3,6-deoxy of beta-D-galactopyranose) would be:
_2(3d)^4P^+1d^a
Note that the +1d remains super-scripted to indicate that the exocyclic, deoxy carbon is ‘up’.
In the case that it is desirable to indicate that the deoxy form definitely came from galactose and not gulose, it would be acceptable to use the following, though it would not commonly be done.
_2^3d,4P^+1d,a
For comparison convenience, beta-D-galactopyranose is: _2^3,4P^+1,a
See also other examples below.
Monosaccharides with Derivatives
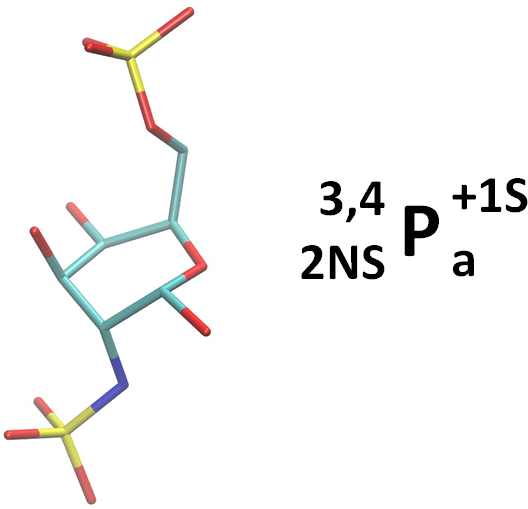
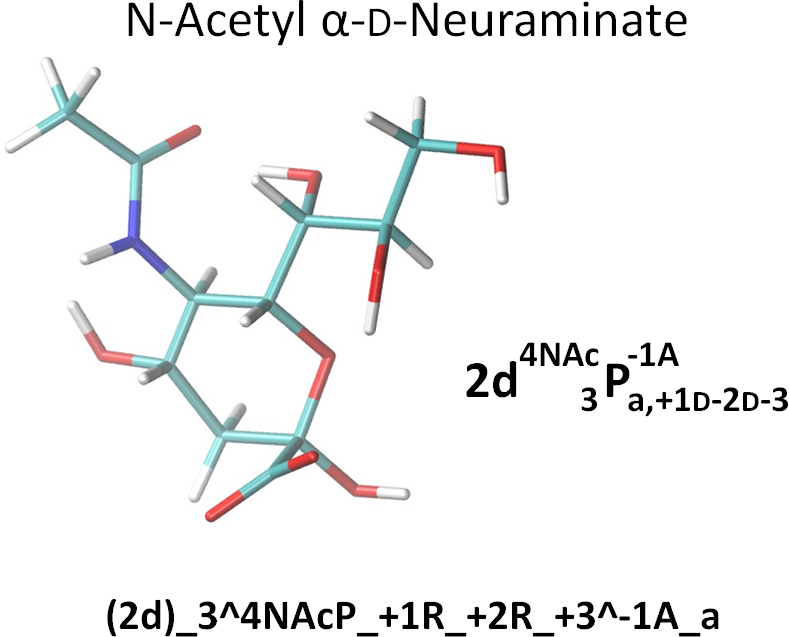
To the extent possible, we use derivative annotations commonly employed in the glycosciences. For example, sulfated positions are marked with ‘S’. And, as per convention, although N-acetylated and N-sulfate positions are also deoxy, we simply use the term ‘NAc’ and ‘NS’. However, if it is necessary to reduce confusion, they could be noted as both ‘d’ (deoxy) and ‘NS’ by ‘d;NS’. Here are two examples. The linearized form is also given in the NeuNAc example.
2-deoxy, 2-N-sulfated, 6-sulfated alpha-D-galactopyranose in a 1C4 ring conformation:
Note that the NeupNAc could also be linearized in a slightly less cumbersome fashion as:
(2d)_3^4NAcP_+{1R-2R-3}^-1A_a
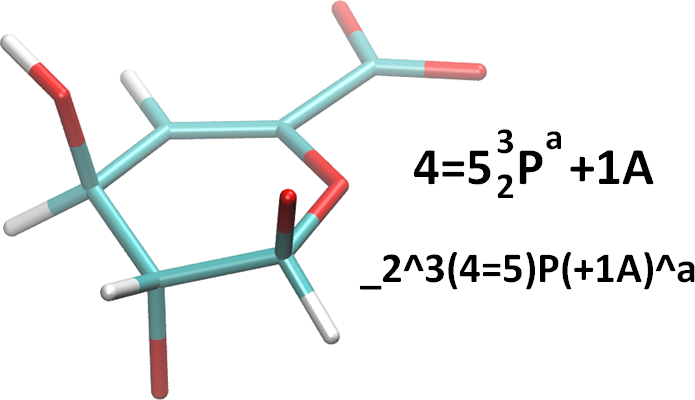
Unsaturated Monosaccharides
It should be stressed that this system was created to aid in recording observed 3D structure. It will rarely be possible to tell conclusively that a double-bond exists in an atomistic structure file such as a PDB file. This will be even less possible if the PDB file does not contain hydrogens. However, in the case that one needs or wishes to indicate the presence of unsaturation, there is a method. It is simple. We use an equals sign (‘=’) between the relevant ring atom numbers. For example, consider the monosaccharide in the figure below. Standard and linearized representations are also shown.
Representing Monosaccharide Linkages
This method is mainly designed to represent monosaccharides. If a large number of monosaccharides were represented in this manner, it would become very difficult for humans to parse. However, there is a method for indicating linkages.
To indicate a linkage location, use a greater-than sign (‘>’) to indicate that the monosaccharide on the left links at the position where the sign is placed. A less-than sign (‘<‘) can be used if it is desirable to indicate that a monosaccharide on the right is connecting. See the example in the figure below.
Note that there need not be any other monosaccharides. For example, to indicate that the alpha-D-glucopyranose used in the examples above is linked to some other (possibly unknown) entities at positions 2, 4 and 6 (corresponding to residue ID RGA in the GLYCAM06 residue naming convention), the standard and linear codes would be:
Standard
>2,>43P>+1a
Linear
_>2^3_>4P^>+1_a
Lookup Tables
Lookup tables are available for the monosaccharides and for names given to commonly-encountered derivatives.
Note that the tables only contain ‘D’ sugars. To generate the ‘L’ sugars, merely swap all the ‘up’ and ‘down’ components so that they are their opposite.
Back Story
This representation started out as Lachele Foley’s personal shorthand for describing monosaccharides. Because she typically provided the technical support for GLYCAM-Web, she often found herself needing to translate a visual representation of a 3D structure into something that could be used to query a lookup table. Having a simple text-based method that could be written down quickly was very useful in this. Oliver Grant named it GlyCode. It is used in this software for the simple reason that Lachele already had lookup tables made.